Katrine Whiteson, PhD
I am interested in understanding how individual and persistent human-associated microbial and viral communities affect health. Infection with a bacterial pathogen, vaccination, immune development and even taking a Tylenol does not occur in a vacuum. Dynamic microbial and viral communities constantly inhabit our bodies, encoding the majority of the unique genes that alter these processes. Resident microbial and viral community composition is unique to each human, with strong similarities across families and also ethnicity.
 Around the time I finished my PhD in biochemistry, high-throughput sequencing technology emerged, and I developed a strong interest in applying microbial ecology approaches to the study of human-associated microbial and viral communities. I took a position at the University of Geneva Hospitals, where I studied human oral microbial communities in healthy people, in response to antibiotic treatment, and in malnourished children who develop gangrenous facial lesions. In my current work in Forest Rohwer's lab at San Diego State University, I focus on the microbial and viral communities inhabiting the lungs of cystic fibrosis (CF) patients. By linking microbial metagenomic sequencing data with volatile metabolites from breath samples (in collaboration with Simone Meinardi and Don Blake at UC Irvine), I aim to identify biomarkers of active microbial physiology to detect infections earlier and treat them more specifically.
Around the time I finished my PhD in biochemistry, high-throughput sequencing technology emerged, and I developed a strong interest in applying microbial ecology approaches to the study of human-associated microbial and viral communities. I took a position at the University of Geneva Hospitals, where I studied human oral microbial communities in healthy people, in response to antibiotic treatment, and in malnourished children who develop gangrenous facial lesions. In my current work in Forest Rohwer's lab at San Diego State University, I focus on the microbial and viral communities inhabiting the lungs of cystic fibrosis (CF) patients. By linking microbial metagenomic sequencing data with volatile metabolites from breath samples (in collaboration with Simone Meinardi and Don Blake at UC Irvine), I aim to identify biomarkers of active microbial physiology to detect infections earlier and treat them more specifically.
I use a combination of omics techniques to characterize the genetic potential of human-associated microbial and viral communities, and connect this with the active chemical processes in the community. Discoveries based on longitudinal and cross-sectional studies of human samples can be tested in model cultures in the lab. The hypothesis underlying my work is that the unique, persistent microbial communities inhabiting the niches that comprise each human undergo recognizable changes in many human disease states. The microbial community structure and metabolic profiles of altered microbial communities are detectable, and can be used to diagnose and monitor infections and lifestyle disease, and tailor treatments to specific microbial states. I have used culture-independent microbial DNA sequencing in my research to demonstrate the stability and persistence of individual oral microbial communities in healthy people, and to examine the origin of a poorly understood infection common in malnourished children (NOMA). Currently, I am monitoring metabolites from Cystic Fibrosis (CF) breath samples and linking them with metagenomic and metatranscriptomic data from paired sputum samples to elucidate the physiology of microbial and viral communities in the CF lung over time.
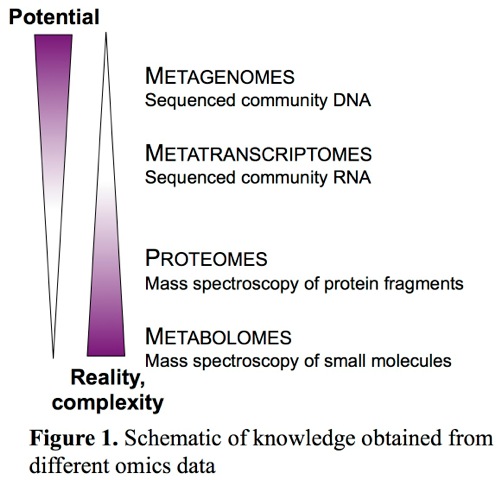 In a sense, metabolomics is an ancient approach. For example, multiple accounts starting around 1500 BC used taste or insects to detect excess glucose in urine and diagnose diabetes. Modern versions of these approaches using dogs or bees to detect conditions such as cancer and tuberculosis all rely on the premise that unique patterns of metabolites emerge from disease. Now we have an array of tools that detect the potential and actual processes in an environment, from high-throughput sequencing of DNA and RNA to metabolite detection with chromatography and mass spectroscopy. A large number of genes and potential metabolic processes come to humans from their resident microbial and viral communities. Metagenomic sequencing provides a profile for this potential, while metatranscriptomes reveal the actively transcribed genes (Figure 1). Finally, metabolomes reveal the products of the chemical reactions occurring in a sample, many of which are unique to a microbial community member or metabolic pathway. In addition to potential identification of an infecting microbe, intercepting chemical communication signals between microbes and the human host is essentially eavesdropping on the system, to learn about the active processes in the community. In combination with well-planned study design, this under-explored territory will yield biomarker signals that can be used to diagnose and monitor health conditions at a greater level of clarity (e.g. earlier diagnosis, or more carefully targeted treatment).
In a sense, metabolomics is an ancient approach. For example, multiple accounts starting around 1500 BC used taste or insects to detect excess glucose in urine and diagnose diabetes. Modern versions of these approaches using dogs or bees to detect conditions such as cancer and tuberculosis all rely on the premise that unique patterns of metabolites emerge from disease. Now we have an array of tools that detect the potential and actual processes in an environment, from high-throughput sequencing of DNA and RNA to metabolite detection with chromatography and mass spectroscopy. A large number of genes and potential metabolic processes come to humans from their resident microbial and viral communities. Metagenomic sequencing provides a profile for this potential, while metatranscriptomes reveal the actively transcribed genes (Figure 1). Finally, metabolomes reveal the products of the chemical reactions occurring in a sample, many of which are unique to a microbial community member or metabolic pathway. In addition to potential identification of an infecting microbe, intercepting chemical communication signals between microbes and the human host is essentially eavesdropping on the system, to learn about the active processes in the community. In combination with well-planned study design, this under-explored territory will yield biomarker signals that can be used to diagnose and monitor health conditions at a greater level of clarity (e.g. earlier diagnosis, or more carefully targeted treatment).
Last modified: Wed Jun 26 12:07:19 PDT 2013
 Around the time I finished my PhD in biochemistry, high-throughput sequencing technology emerged, and I developed a strong interest in applying microbial ecology approaches to the study of human-associated microbial and viral communities. I took a position at the University of Geneva Hospitals, where I studied human oral microbial communities in healthy people, in response to antibiotic treatment, and in malnourished children who develop gangrenous facial lesions. In my current work in Forest Rohwer's lab at San Diego State University, I focus on the microbial and viral communities inhabiting the lungs of cystic fibrosis (CF) patients. By linking microbial metagenomic sequencing data with volatile metabolites from breath samples (in collaboration with Simone Meinardi and Don Blake at UC Irvine), I aim to identify biomarkers of active microbial physiology to detect infections earlier and treat them more specifically.
Around the time I finished my PhD in biochemistry, high-throughput sequencing technology emerged, and I developed a strong interest in applying microbial ecology approaches to the study of human-associated microbial and viral communities. I took a position at the University of Geneva Hospitals, where I studied human oral microbial communities in healthy people, in response to antibiotic treatment, and in malnourished children who develop gangrenous facial lesions. In my current work in Forest Rohwer's lab at San Diego State University, I focus on the microbial and viral communities inhabiting the lungs of cystic fibrosis (CF) patients. By linking microbial metagenomic sequencing data with volatile metabolites from breath samples (in collaboration with Simone Meinardi and Don Blake at UC Irvine), I aim to identify biomarkers of active microbial physiology to detect infections earlier and treat them more specifically.

 In a sense, metabolomics is an ancient approach. For example, multiple accounts starting around 1500 BC used taste or insects to detect excess glucose in urine and diagnose diabetes. Modern versions of these approaches using dogs or bees to detect conditions such as cancer and tuberculosis all rely on the premise that unique patterns of metabolites emerge from disease. Now we have an array of tools that detect the potential and actual processes in an environment, from high-throughput sequencing of DNA and RNA to metabolite detection with chromatography and mass spectroscopy. A large number of genes and potential metabolic processes come to humans from their resident microbial and viral communities. Metagenomic sequencing provides a profile for this potential, while metatranscriptomes reveal the actively transcribed genes (Figure 1). Finally, metabolomes reveal the products of the chemical reactions occurring in a sample, many of which are unique to a microbial community member or metabolic pathway. In addition to potential identification of an infecting microbe, intercepting chemical communication signals between microbes and the human host is essentially eavesdropping on the system, to learn about the active processes in the community. In combination with well-planned study design, this under-explored territory will yield biomarker signals that can be used to diagnose and monitor health conditions at a greater level of clarity (e.g. earlier diagnosis, or more carefully targeted treatment).
In a sense, metabolomics is an ancient approach. For example, multiple accounts starting around 1500 BC used taste or insects to detect excess glucose in urine and diagnose diabetes. Modern versions of these approaches using dogs or bees to detect conditions such as cancer and tuberculosis all rely on the premise that unique patterns of metabolites emerge from disease. Now we have an array of tools that detect the potential and actual processes in an environment, from high-throughput sequencing of DNA and RNA to metabolite detection with chromatography and mass spectroscopy. A large number of genes and potential metabolic processes come to humans from their resident microbial and viral communities. Metagenomic sequencing provides a profile for this potential, while metatranscriptomes reveal the actively transcribed genes (Figure 1). Finally, metabolomes reveal the products of the chemical reactions occurring in a sample, many of which are unique to a microbial community member or metabolic pathway. In addition to potential identification of an infecting microbe, intercepting chemical communication signals between microbes and the human host is essentially eavesdropping on the system, to learn about the active processes in the community. In combination with well-planned study design, this under-explored territory will yield biomarker signals that can be used to diagnose and monitor health conditions at a greater level of clarity (e.g. earlier diagnosis, or more carefully targeted treatment).